By Vishwanath Prathikanti, Anthropology, ‘23
Author’s note: As someone studying Anthropology at Davis, I often see my friends confused when I tell them how much of my studies consist of biology and chemistry. It’s a fairly common conception that Anthropologists mainly study human culture, and while cultural anthropology is an important aspect of the field, it is still only a part of it. When I heard about how our ancestors’ enzymes are being used to advance our knowledge of cancer, I knew it could be an opportunity to change the perception of Anthropology among students.
Most people have a general understanding of how cancer works: it occurs when apoptosis, or cell death, does not occur in cells. These cells start to propagate, and then aggregate into tumors. The tumors can spread across the body and lead to varying health complications depending on if they are benign (isolated to a part of the body) or malignant (spread to other areas). Naturally, one possible solution would be to fix the part in cancer cells that prevent them from properly dying. So how does a cell die?
Apoptosis hinges on enzymes called effector caspases, which deactivate proteins that carry out normal cellular processes, activate nucleases and kinases that are used to break down DNA, and disassemble various components of the cell [1]. So to cause cell death in cancer cells, scientists would need to activate caspases. Activating these caspases would affect all cells, not just cancerous ones. The challenge scientists face is activating caspases in cancer cells without impacting healthy surrounding cells. Unfortunately, to activate effector caspases in just cancerous cells requires an intimate knowledge of the different proteins that comprise the caspase family, something the scientific community lacks.
In an effort to learn more about the structure of caspases, Suman Shrestha and Allan C. Clark from the University of Texas at Arlington decided to look to the past rather than just the present. Specifically, they wanted to analyze the folding mechanisms and structure of effector caspases and construct a picture of how they operated for our ancestors [2].
A recent trend in evolutionary biology and physical anthropology has been comparing various proteins and their folding structures across other organisms today and reconstructing what these proteins looked like for our ancestors [3]. This is carried out via a computer program that generates a phylogenetic tree of a protein family, a process known as ancestral sequence reconstruction (ASR). After the phylogeny is generated, the ASR program will statistically infer where certain proteins changed or emerged in the tree [4]. This is done by comparing binding sites in proteins. The program will identify various binding sites that are described as “ambiguous sites,” when a node (branching point in a phylogenetic tree) has a <70% probability of being accurate [5]. In caspases, this ambiguity is generally due to one of two possibilities. One is that there is nearly a 50/50 chance an identified ancestral protein led to the extant version, or another identified protein. The second possibility is that the binding site has a high mutation rate, lowering the probability that it has been characterized correctly [5]. As for the other sites, different ASR programs have slightly different levels of accuracy, but the most prominently used ones have around a 90-93% chance that every non-ambiguous site is accurate [8]. Finally, using protein sequences of the organisms alive today and the phylogeny that depicts their ancestors, the ASR program can present the most likely sequence of the protein at a particular node in the phylogeny [6].
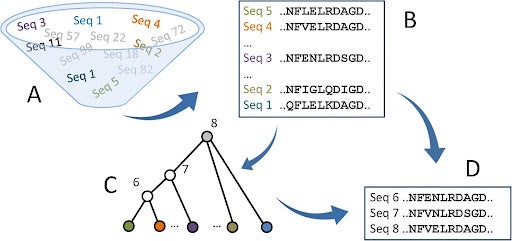
Caption: The ASR process will generate the phylogeny (C) as well as the sequences and order of sequences provided those of extant species are provided to the program (D) [4].
Using ASR, Shrestha and Clark discovered effector caspases first evolved in a common ancestor more than 650 million years ago (mya) when microorganisms and sponges dominated life. While ASR can’t identify the species of the organism, it can generate the predicted sequences of these ancient caspases. This is all they need to recreate these proteins and better understand how these caspases function under healthy conditions versus cancerous ones [2, 7].
Among the 12 proteins that make up the caspase family, Shrestha and Clark decided to reconstruct the ancestor of three specific ones: caspase-3, -6, and -7 [7]. These three caspases were chosen because they are specifically responsible for cell death, whereas the others are linked to inflammation or activation of other enzymes [7, 8]. After sequencing the proteins, Shrestha and Clark were able to identify changes in the folding structures and sequences that could activate effector caspases in tumor cells without triggering cell death in healthy cells.
Specifically, they confirmed two folding precursors in the creation of caspase-6 and -7 proteins. While these precursors had already been discovered in caspase-3, the discovery was significant in understanding how the caspases worked in a normal cell and how they were altered in a cancer cell. Shrestha and Clark noted mutations that slow the formation of these precursors, which led to the production of caspases greatly slowing down, causing a cell to not die when it needs to [2]. Understanding this regulatory process may allow researchers to discover a way to reactivate caspase production in cancer cells.
The vast majority of data collected in the study was information on how stable these proteins are and where they evolved since our common ancestor 650 mya. They found that caspase-6 was the most stable out of the three, and at lower pH’s, caspase-6 is the only one that does not unfold irreversibly [2]. This suggests a more specialized role for caspase-6 compared to 3 and 7, and the data may be useful for the adaptation of cancer-targeting drugs. For example, if a cancer aggregate is in a low pH environment of the body such as the stomach, a cancer-targeting drug may utilize caspase-6 specifically to activate programmed cell death.
While the results are still fairly recent and have not had adequate time to be implemented into a treatment, Morteza Khaledi, dean of the College of Science at the University of Texas at Arlington, was incredibly excited about the results. In a press statement to the University of Texas at Arlington, he announced that the research had yielded “vital information about the essential building blocks for healthy human bodies” and that the knowledge gained from the study will be seen as “another weapon in our fight against cancer” [7].
References:
- https://www.sciencedirect.com/topics/medicine-and-dentistry/effector-caspase
- https://www.sciencedirect.com/science/article/pii/S0021925821010528?via%3Dihub
- https://www.nature.com/articles/nrg3540
- https://onlinelibrary.wiley.com/doi/10.1002/bip.23086
- https://www.sciencedaily.com/releases/2022/01/220112094022.htm
- https://link.springer.com/chapter/10.1007/978-1-4614-3229-6_4?utm_source=getftr&utm_medium=getftr&utm_campaign=getftr_pilot
- https://portlandpress.com/biochemj/article/476/22/3475/221018/Resurrection-of-ancestral-effector-caspases
- https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.0020069#:~:text=The%20ML%20method%20was%20the,average%20accuracy%20is%200.4%25
- https://www.sciencedirect.com/science/article/pii/S2213020916302336